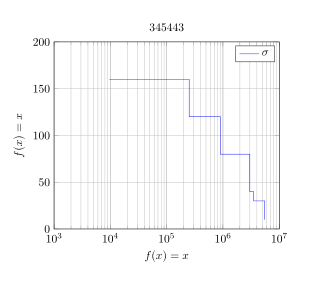
|
Wir ihr bei meinem Testbeispiel seht, ist meine x-Achse logarithmisch jedoch aber die y-Achse linear. Die Daten lese ich in Form einer .dat Datei ein. Anbei meine .dat Datei: spalteB spalteA 160 9600 120 251000 80 900000 40 3000000 30 3500000 10 5500000 Anbei mein Testbeispiel: \documentclass{article} \usepackage{pgfplots} \begin{document} \begin{tikzpicture} \begin{semilogxaxis} [ enlarge x limits=false, no marks, grid=both, xmin=0, xmax=1e7, ymin=0, ymax=200, legend entries={$\sigma$} title=345443, ylabel={$f(x)=x$}, xlabel={$f(x)=x$} ] %\addplot+[const plot] \addplot+ table[x=spalteA,y=spalteB, const plot] {1.dat}; \end{semilogxaxis} \end{tikzpicture} \end{document} Was ich gerne erreichen würde:
Ein Stufendiagramm ohne manuelle Berechnung der "Zwischen Punkte".
|
|
Du musst \addplot+[const plot] table[x=spalteA,y=spalteB] {1.dat};
Code: \documentclass{article} \usepackage{pgfplots} \pgfplotsset{compat=1.14} \begin{document} \begin{tikzpicture} \begin{semilogxaxis} [ enlarge x limits=false, no marks, grid=both, xmin=1e3, xmax=1e7,% <- xmin auf sinnvollen Wert geändert ymin=0, ymax=200, legend entries={$\sigma$},% <- hier fehlte das Komma title=345443, ylabel={$f(x)=x$}, xlabel={$f(x)=x$} ] \addplot+[const plot] table[x=spalteA,y=spalteB] {1.dat}; \end{semilogxaxis} \end{tikzpicture} \end{document} |